How Formaspace Helped Sequence the Human Genome

Steven Riedmuller, who was Senior Director of Field Applications at TIGR at the time
When Craig Venter’s team was sequencing the first human genome, TIGR relied on FORMASPACE to give them a competitive edge over publicly-funded contesting labs.
We now consider it normal to collect DNA samples as evidence at a crime scene to implicate (or exonerate) suspects. Taking this further, residents of upscale New York City co-ops have even gone as far as testing doggie poo to nab the dog owners who don’t clean up after their pets.
We can take a swab sample (often in the comfort of our own home) to determine which viruses we have been exposed to during our lifetime, identify diseases which our individual genes might make us more likely to develop, or get insight into our ancestral genetic racial heritage. We can even get our own dogs tested to determine their breed backgrounds.
Doctors can now provide personalized medicine (pharmacogenomics) at leading healthcare institutions, like MD Anderson Cancer Center, which tests patients for their unique genetic makeup in order to provide ‘precision medicine’ cancer treatments. Eventually, the “Moon Shot”, or the cure-all for cancer, could also be readily available.
Public health officials and investigators can identify strain variations of disease pathogens, ranging from MRSA and HIV to Anthrax, making it possible in many cases to determine the source of a disease outbreak.
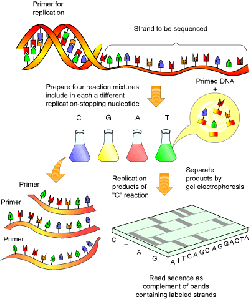
How did laboratory scientists break the code of the Human Genome?
Let’s roll the clock back to the immediate post-World War II period. Cambridge University graduate Frederick Sanger began his career studying insulin protein at the National Institute for Medical Research in London. This work led the Nobel Committee to award Sanger a Nobel Prize for successfully sequencing insulin and determining conclusively that amino acid proteins had a defined, regular sequence.
While Sanger had been studying insulin in London, British scientist Francis Crick and American researcher James D. Watson proposed a novel solution to explain the structure of DNA — two strands of DNA coiled together to form a double helix.
Importantly, they theorized that genetic information flowed in one direction — from DNA to RNA, then from RNA to proteins. (Previously many scientists, including Sanger, thought proteins — not DNA — were the source of genetic information.)
An experiment comparing regular hemoglobin with hemoglobin affected by sickle-cell anemia confirmed the new DNA replication hypothesis.
Sanger eventually joined Crick and Watson at Cambridge, where he studied how DNA makes proteins using messenger RNA (mRNA) and transfer RNA (tRNA) at the new Laboratory of Molecular Biology (LMB).
Meanwhile other researchers around the world were making progress. Robert Holley’s team at Cornell was the first the sequence alanine transfer RNA (tRNA) from yeast; it contained 77 nucleotides.
The first DNA sequencing breakthrough came from researchers Wu and Kaiser, who used DNA polymerase to sequence a small fragment of lambda bacteriophage DNA; making it the first time DNA had been sequenced.
The number of identified DNA sequences began to grow and grow.
By 1977, Sanger identified the first complete genome of an organism, the phi X 174 bacteriophage, containing 5,386 nucleotides.
Sanger and Coulson published their gene sequencing technique in 1977 — today known as “Sanger’s Method” — which uses dideoxynucleotides to control the DNA polymerase reactions.
Meanwhile, Allan Maxam and Walter Gilbert had developed their own technique that could sequence double-stranded DNA, making it a superior method despite the use of highly toxic chemicals.
However, in the end, Sanger’s Method became the dominant first-generation sequencing technique when Bruno Gronenborn and Joachim Messing demonstrated they could insert a double-stranded DNA into a single-stranded M13 bacteriophage. This bacteriophage would then ‘clone’ the DNA sequence into single strands of DNA – making it was suitable for analysis using the Sanger’s Method.
For their scientific research, Maxam, Gilbert and Sanger were awarded the 1980 Nobel Prize in Chemistry, making Sanger a rare two-time winner. In 1983 Frederick Sanger retired; he passed away in 2013.
(Bacteriophage viruses are in the news lately. They are the ‘knives’ used to cut DNA in the CRISPR/Cas9 technique we have written about previously.)
The Rise of Automation Techniques for Gene Sequencing
As the scope of DNA sequencing projects grew, it became imperative to develop computer software and laboratory automation systems to process and interpret the vast amount of data. By the late 1970s, researchers like Roger Staden at the LMB were programming minicomputers to manipulate scientific data.
In the late 1980s, the Caltech scientists introduced the AB370, the first automated DNA sequencer. It used fluorescent dyes to identify DNA bases, rather than radioactive tagging used in the classic Sanger’s Method. Soon the new DNA sequencers were fast enough to sequence 50,000 bases in a few hours – previously the same task took a full man-year to complete.
The Start of the Human Genome Project
With the successful completion of a three year project to sequence human mitochondrial DNA, it was time to tackle the biggest objective of all: sequencing the entire human DNA.
The Human Genome Project was launched in 1990 with a budget of $3 billion, with participants from science labs around the world.
But it turns out they were not the only ones wanting a crack at the problem. In 1992, American researcher J. Craig Venter left the National Institutes of Health to establish his own research unit called The Institute for Genomic Research (TIGR).
In 1998, Venter established another new venture called Celera Genomics, whose goal was to sequence the human genome faster than the public-funded Human Genome Project — using a method called whole genome “shotgun” sequencing.
We spoke with Steven Riedmuller, who was Senior Director of Field Applications at TIGR at the time.
During Steven Riedmuller’s tenure as the Laboratory Automation Manager at TIGR, the race between Venter’s team and The Human Genome Project was coming down to the wire. Which team would decode the Human Genome first?
In response to the challenge, Venter’s organization went big: they decided they needed to double their genetic sequencing capacity as soon as physically possible.
But despite the herculean effort, the TIGR facility had a physical capacity problem — one that many laboratory managers face every day. How could they fit double the amount of lab equipment (in this case DNA sequencers) when there simply wasn’t enough space in the existing lab?
The easy answer, facility expansion, was out of the question. There wasn’t time to lease another building, much less equip it. And the cost for that was going to be a budget buster.
That’s when TIGR’s Laboratory Automation Manager, Steven Riedmuller, called Formaspace to see how we could help them out with an innovative solution.
What we discovered is that weight and cost were two important issues. The sequencers at TIGR were heavy (weighing in at 400 pounds each) and valuable — each sequencer unit cost around $140,000 each!
Here at Formaspace, we knew even our standard workbenches were strong enough to ensure that these heavy, expensive sequencers were secure and stable. The trick was to find a way to increase the capacity of the TIGR laboratory without increasing their square footage footprint.
The solution? We fabricated heavy-duty modular workbenches that nested together in a double stack – allowing TIGR to use twice the number of sequencers in their existing laboratory footprint.
The result: TIGR doubled their DNA sequencing productivity rate — and, very importantly — they avoided having to fork over $5 million dollars on a time consuming real-estate expansion.
Time for required for Formaspace to fabricate and deliver? A mere 8 weeks from start to finish.
Read More. https://formaspace.com/articles/laboratory-furniture/formaspace-helped-sequence-human-genome/?utm_source=einpresswire&utm_medium=content&utm_campaign=article-06072016
Mehmet Atesoglu
Formaspace
8002511505
email us here
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.